By Amna Qamar, M. Zulqurnain Imran, Nayab Afzal, Shabbir Hussain, Madiha Andleeb, Noureen Shakeel
AFFLIATIONS:
Department of Pathology, National Medical Centre, Karachi, Pakistan.
DOI: https://doi.org/10.36283/PJMD11-2/005
How to cite: Qamar A, Imran MZ, Afzal N, Hussain S, Andleeb M, Shakeel N. Frequency of BCR-ABL Gene Translocation in B-ALL Patients Associated with Clinicopathological Parameters. Pak J Med Dent. 2022;11(2): 22-28. doi: 10.36283/PJMD11-2/005
Background: B-Acute Lymphoblastic Leukemia (B-ALL) accounts for 25% of childhood malignancies. Chromosomal abnormalities like translocations lead to the formation of oncogenes, some of which are strong predictors of prognosis and response to anti-leukemic therapy. This study aimed to find out the frequency of BCR-ABL gene translocation in B-ALL patients by fluorescence in situ hybridization (FISH) and its association with their clinicopathological parameters.
Methods: Patients (n=150) aged 1-17 years with a confirmed diagnosis of B-ALL were selected. Peripheral blood and/or bone marrow aspirate samples were obtained and Breakpoint cluster region-Abelson murine leukemia viral oncogene (BCR-ABL) translocation by FISH was observed. The patient’s demographics, hemoglobin levels, total leucocytes count, platelet count, FISH results, CNS status and risk stratification were recorded. Data was stratified and the Chi-square test was applied, p-value < 0.05 was considered statistically significant.
Results: There were 100 (66.67%) males and 50 (33.33%) females. The average age of the patients was 7.03±4.51 years. The frequency of BCR-ABL translocation in B-ALL was 16(10%). A significant association was found between age and BCR-ABL translocation (p-value <0.05), however, an insignificant association was recorded among gender, Hb levels, TLC, platelet count, CNS status, proposed risk stratification system of B-ALL and BCR-ABL translocation (p-value >0.05).
Conclusion: The frequency of BCR-ABL translocation in B-ALL was significantly high in the targeted population. FISH improves detection of the BCR-ABL translocation in either metaphase or interphase cells. Therefore, BCR-ABL expression can be considered as a prognostic approach and assist in effective treatment planning and better management of these children.
Keyword: Leukemia; In Situ Hybridization; Fluorescence; Pathology.
Acute lymphoblastic leukemia (ALL) is one of the most commonly occurring malignancies among children under 15 years of age causing about 25% of the childhood malignancies1. Acute leukemias are frequently associated with various chromosomal disorders like translocations, deletions, etc., many of these translocations result in the formation of abnormal fusion genes which are called oncogenes. It has been noted that a number of these translocations found in ALL patients act as strong prognostic predictors and play a significant role in as response of cells to antileukemic therapy. Reciprocal translocations were initially thought to occur in only chronic myelogenous leukemia (CML), but now it is a well-known fact that such translocations are also found in acute leukemia. One very important reciprocal translocation of BCR-ABL has been identified in acute leukemia and other hematopoietic disorders in both adults and children2-4.
BCR-ABL translocation as shown in Figure 1 refers to the juxtapositioning of the ABL-1 gene involving the chromosome 9 (q:34) to the BCR gene on chromosome 22(q:11). This produces Philadelphia chromosome p190 BCR-ABL1(e1a2) constitutes the BCR-ABL1 transcript (m-BCR). This new chromosome encodes a hybrid protein called 190-kDa protein. p190BCR-ABL is commonly detected in B-ALL4-6.
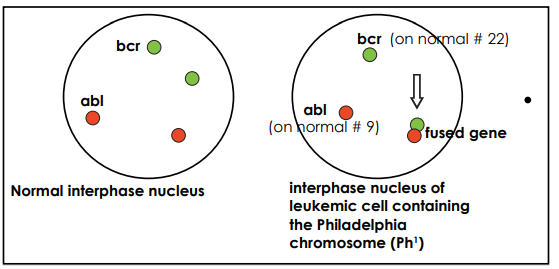
Figure 1: Fluorescence in situ hybridization detection of breakpoint cluster region protein and the Abelson murine leukemia (BCR-ABL).
Studies conducted earlier have concluded that Philadelphia (Ph) chromosome is found in about 20% to 30% of adult acute lymphoblastic leukemia (ALL) and 5% of ALLs affecting children7,8. BCR-ABL translocation can be detected by conventional karyotyping methods. Conventional karyotyping detects most BCR-ABL related translocations with the identification of its different partner chromosomes. Fluorescence in situ hybridization (FISH) uses a dual-color dual fusion probe. It is a useful technique to detect BCR-ABL translocation in interphase cells in a rapid and sensitive manner9. The presence of BCR-ABL translocation in ALL is associated with aggressive disease and has been shown to have a poor prognosis, especially in children. Thus, such patients are candidates for more aggressive treatment regimens with tyrosine kinase inhibitors10. This study was designed to find out the frequency of BCR-ABL gene translocation in B-ALL children and its association with their demographic and clinicopathological parameters.
This cross-sectional descriptive study was conducted from 1st August 2020 to 28th February 2021. All 150 cases were selected through the non-probability purposive sampling technique.ERC has approved the study with the reference code (Reference code: 2841120AIHEM). All newly diagnosed B-ALL patients, aged 1-17 years with either gender were included in the study. The researchers included ALL cases while atypical cases and other types of leukemia like Acute Myeloid Leukemia, T-Acute Lymphoblastic Leukemia, and Chronic Leukemia were not included in the study. Hb levels, TLC, platelet count were recorded. TLC levels were categorized into following groups, <5×109/l, 5-50 x109/l and >50 x109/l. Hemoglobin levels were classified into two groups of <8 gm/dl and >8 gm/dl. Platelets were divided into three groups as follows, <50 x109/L, 50-100 x109/L and >100 x109/L. All the samples were analyzed on an automated hematology analyzer, Sysmex XN 350.
Study protocol and purpose were explained to the patients’ attendant and consent was taken. Demographic data i.e., age, gender was entered into predesigned proforma. Peripheral blood and/or bone marrow aspirate was obtained in a sodium heparin vacutainer tube. Slides were prepared in cytospin and slides were kept for overnight aging. The following day slides were washed in formamide, dehydrated with alcohol and dried. In the next step dual color, dual fusion translocation probe was added and slides were kept in ThermoBrite for hybridization. On the 3rd day, post hybridization slides were washed, the counterstain was added and slides were kept in dark. Prepared slides were observed in an immunofluorescence microscope using filter DAPI (green red) and DAPI (green orange). For each sample, 200 non-overlapping cells were counted. All the normal cells (in which there is no translocation 9;22) exhibited two separate red and two separate green signals. On the other hand, cells containing a simple balanced t(9;22) translocation, the typical pattern as one red, one green signal from the normal 9 and 22 chromosomes and two red/green (yellow) fusion signals, one each from the derivatives 9 and 22 were observed as shown in Figure 2. The cut-off percentage for typical t(9;22) was taken as 0.5%. All the children were assessed for their CNS status.
CNS1: in the cerebral spinal fluid (CSF), absence of blasts on cytospin preparation, irrespective of the number of WBCs, CNS2: in CSF, presence <5/ul WBCs and cytospin positive for blasts, or traumatic LP, >5/ul WBCs. CNS 3: CSF, after traumatic LP presence of > 5/ul WBCs and cytospin positive for blasts and/or clinical signs of CNS leukemia. Patients were classified into standards risk (age >1year to <10year, TLC<50,000/mm3, Not Tel-AML or Trisomy 4,10,7), high risk (Age >10-year, TLC>50,000/mm3, CNS3 or testicular disease, MLL translocation) and very high-risk groups (Philadelphia positive leukemia, hypodiploidy >44 chromosomes, induction failure) at time of diagnosis. Data were analyzed in SPSS version 22. Mean±SD was calculated for parametric data. Median and interquartile range was calculated for non-parametric data like age. For categorical variables like gender, CNS status, risk and outcome variables i.e., BCR–ABL positivity or BCR-ABL negativity. A Chi-square test was applied, p-value <0.05 was considered as statistically significant.
A total of 150 diagnosed cases of B-lymphoblastic leukemia meeting the inclusion criteria were included in this study. There were 100(66.67%) males and 50(33.33%) females with the age ranging from 01 to 17 years. The average age of the patients was 7.03±4.51 years (95% CI:6.29 to 7.76). And median TLC count was 29.75/mm3 (IQR=61.53), median platelet count was 72.00(IQR=66.25), mean Hb was 8.67+1.73 gm/dl.
The study subjects were divided into four groups i.e., <1 year, 1-10 and >10 years. Most patients were in the 1–10 years age group, 110(73.3%). The frequency of BCR-ABL translocation by FISH in 1-17 years patients with B-ALL was 15 (10%). Patients were analyzed concerning different variables including age, gender, Hb levels, TLC, platelet count, CNS status and risk stratification. The rate of BCR-ABL translocation was significantly high among the individuals of age group >10 years (p=0.001) as shown in Table 1.
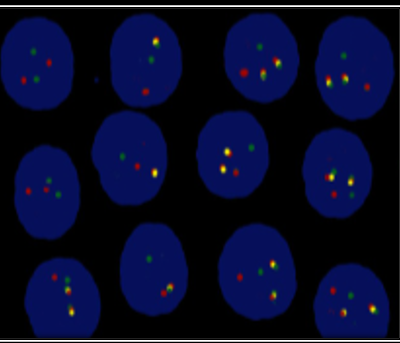
Figure 2: Fluorescence in situ hybridization detection of breakpoint cluster region protein and the Abelson murine leukemia (BCR-ABL) and abnormal fusion gene highlighted as yellow signals.
The rate of BCR-ABL translocation was not significant between males and females (p =1.00). For risk stratification at the time of diagnosis; 63(42%) were classified as standard risk, and 2 (1.3%) were classified as very high risk as per leukemia risk stratification protocol. p-value concerning CNS status and risk stratification was found to be 0.348 and 0.373 respectively. No significant association was found between TLC, Hb levels, platelet count, p-value 0.239, 0.883 and 0.168 respectively as shown in Table1.
Table 1: Frequency of detection of BCR-ABL fusion by FISH in B-ALL children concerning age gender, CNS status risk stratification, TLC, Hb and platelet count.
| Variables | Frequency (n) (%) | BCR-ABL Fusion | p-Value | |
| Positive
(n) (%) |
Negative
(n) (%) |
|||
| Age Groups (Years) | ||||
| < 1Years | 3(2%) | 0(0%) | 3(100%) | 0.001 |
| 1 to 10Years | 110(73.4%) | 5(4.5%) | 105(95.5%) | |
| >10 Years | 37(24.6%) | 10(27%) | 27(73%) | |
| Gender | ||||
| Male | 100(66.7%) | 10(10%) | 90(90%) | 1.00 |
| Female | 50(33.3%) | 5(10%) | 45(90%) | |
| CNS Status | ||||
| CNS-1 | 95(63.33%) | 9(9.5%) | 86(90.5%) | 0.348 |
| CNS-2 | 43(28.6%) | 6(14%) | 37(86%) | |
| CNS-3 | 12(8.6%) | 0(0%) | 12(100%) | |
| Risk | ||||
| Standard | 63(42%) | 4(6.3%) | 59(93.7%) | 0.373 |
| High | 85(56.6%) | 11(12.9%) | 74(87.1%) | |
| Very High | 2(1.3%) | 0(0%) | 2(100%) | |
| TLC | ||||
| <5 x109/l | 21(14%) | 2(9.5%) | 19(90.5%) | 0.239 |
| 5-50×109/l | 73(48.7%) | 5(6.8%) | 68(93.2%) | |
| >50×109/l | 56(37.3%) | 9(16. 1%) | 47(83.9%) | |
| Hemoglobin | ||||
| <8gm/dl | 50 (33.3%) | 6(12%) | 44(88%) | 0.883 |
| >8gm/dl | 100(66.7%) | 11(11%) | 89(89%) | |
| Platelets | ||||
| <50 x109/L | 58(38.6%) | 5(8.6%) | 53(91.4%) | 0.168 |
| 50-100×109/L | 55(36.7%) | 4(7.3%) | 51(92.7%) | |
| >100×109/L | 37(24.6%) | 7(18.9%) | 30(81.1%) | |
BCR-ABL= breakpoint cluster region protein and the Abelson murine leukemia, FISH= fluorescence in situ hybridization, B-ALL= B-cell acute lymphoblastic leukemia, CNS=central nervous system, TLC=total leukocyte count, Hb= hemoglobin.
B-Lymphoblastic Leukemia (B-ALL) is a common childhood malignancy in Pakistan. It affects T-cells and B-cells of the immune system. In children usually, B-cells are the main involved cell line. It is a genetic disease with the involvement of many fusion-related oncogenes. These fusion oncogenes have great prognostic significance and determine the type of appropriate therapy and patient outcome. BCR-ABL translocation also referred to as Philadelphia Chromosome is seen in about 02 to 03% cases of B-ALL across the globe11. The Philadelphia chromosome or Philadelphia translocation (Ph chromosome) results from a reciprocal translocation between the long arms of chromosomes 9 and 22 and can be found in all hematopoietic precursors cells. This translocation results in information of a new fusion protein that can be seen in malignant hematopoietic cells12. In B-ALL, the Ph chromosome disrupts the physiological signaling but also genomic stability of the hematopoietic cells13.
This translocation results in rapid cell proliferation, resistance to apoptosis and cell death.14,15. Translocation causes many changes at the epigenetic level, abnormalities and mutations called copy number variations resulting in the very aggressive clinical course. The presence of the BCR-ABL gene results in more DNA double-strand breaks. After this DNA breakage usually, the repairs are imperfect result in the accumulation of mutations in the genes. These mutations cause drug resistance and rapid disease progesssion16,17. The overall clinical outcome of B-ALL in children is majorly determined by the type and dose of chemotherapy used. Special drug regimes with doses adjusted according to pediatric needs have been designed. The prognosis of B-ALL patients who have BCR-ABL translocation is very poor if treated with standard chemotherapy18. This poor prognosis has led to the early adoption of modified treatment options importantly tyrosine kinase inhibitors (TKIs).
B- lymphoblastic leukemia is a very common malignancy of childhood but there is a paucity of data regarding the frequency of this translocation in the population. A deeper knowledge could help us in decision-making about the appropriate treatment of such patients. The pediatric population from various regions of Pakistan who visited the facility was included in this study. The current study showed that the prevalence of BCR-ABL translocation in the set of individuals was 10% which is less than that reported in other studies from Pakistan i.e., Awan et al. and Faiz et al, however, Siddiqui et al. reported a much lower prevalence of BCR-ABL translocation in a pediatric population. International data have reported similar or much low prevalence18-20. Pandita showed higher results than the present study. This difference could be explained based on variation in sample size, race, and ethnicity21. The mean Hb was 8.67+1.73 gm/dl, this means that most of the children with B-ALL are found to be anemic. Similar findings have been reported by Siddiqui et al20. However, no significant difference was found between Hb level, WBC count and platelet counts in BCR-ABL fusion-positive and negative patients. Likewise, has been reported by Schlieben et al22. Most individuals (73%) who were positive for BCR-ABL were above 10 years of age which is like other local studies. There was a statistically significant association between age and BCR-ABL positivity (p=0.001) pointing towards a worse prognosis in older children if TKIs are not used in the treatment plan. Subsequently, age can be regarded as an independent prognostic factor for the prognosis of B-ALL. However, another researcher has reported otherwise23. No significant association was found between gender and risk stratification (p>0.05) in the current study.
These findings suggest that prognosis does not differ much concerning gender, or risk stratification at the time of diagnosis. CNS involvement was assessed through CSF examination after lumbar puncture. No clinically significant association was found between CNS involvement and BCR-ABL positivity in children included in the study (p=0.348). Hence irrespective of the CNS involvement all the children should be considered for the TKI treatment regime, to protect against CNS infiltration. Other studies have also noted a low CNS involvement at the time of initial diagnosis and higher prevalence at the time of relapse22. There was no independent prognostic impact of different cell counts. These findings are following another study by Cao et al 24.
Results of this study showed that age is the most important factor while treating a child with B-ALL. All the children with a diagnosis of B-ALL must be screened for BCR-ABL translocation, to achieve better treatment outcomes and avoid the toxic effects of other chemotherapy protocols. A significant finding of the study which is comparable to other local and international studies shows a rising frequency of BCR-ABL positivity in childhood B-ALL for age. We observed that the frequency of BCR-ABL translocation in B-lymphoblastic leukemia found in the set of individuals is more than most of the overall incidences as reported in the international data in which the prevalence is mostly low as compared to the study data as shown in Table 2.
Table 2: Comparison of the current study with studies of pediatric acute lymphocytic leukemia (ALL) conducted internationally.
| Studies | Pediatric Population | Reported Frequency of BCR-ABL in ALL |
| Present Study | 1-17 years | 10% |
| Artigas et al. 25 | 1month-14years | 4% |
| Pandita et al. 21 | 1-19 years | 15% |
| Trka et al. 26 | 1-18 years | 2.5 |
| Siraj et al. 27 | <1-21 years | 5% |
| Bekker-Méndez et al. 28 | <19 years | 1.1% |
BCR-ABL= breakpoint cluster region protein and the Abelson murine leukemia.
This study has a few limitations. Firstly, this was a single-center study. Secondly, the sample size could have been larger and lastly patients were not followed up for the kind of treatment offered and their outcomes. Consequently, further large-scale, multicenter studies are warranted for a better understanding of the true reflection of this particular entity and the impact of the modified treatment approach on outcome.
The frequency of BCR-ABL translocation in B-lymphoblastic leukemia found in the targeted population was 10%, as international data. It has a significant association with increasing age. There is a need for earlier registries, investigations and collaboration throughout different parts of Pakistan to make treatment of this disease a possibility.
The authors are grateful to technologists at the Hematology Department for their facilitation in sample processing and analysis during this study.
Authors have no conflict of interest among them.
Organization has approved the study with the reference code (Reference code: 2841120AIHEM).
Informed consent was taken from parents/guardians of all participants. Anyone wishing to discontinue being part of the study was respected and done so.
AQ supervised the research, wrote the manuscript. ZI proposed the research question. NA analyzed the data, assisted the corresponding author. SH maintained data transparency, MA contribution in fluorescence in situ hybridization (FISH) images and NS performed the data entry.
- Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, et al. SEER cancer statistics review, 1975–2010. Bethesda (MD): National Cancer Institute. Based on November 2012 SEER data submission, posted to the SEER website, 2013. Available from: https://seer.cancer.gov/csr/1975_2017.
- Qin Z, Xiang C, Zhong F, Liu Y, Dong Q, Li K, et al. Transketolase (TKT) activity and nuclear localization promote hepatocellular carcinoma in a metabolic and a non-metabolic manner. J Exp Clin Cancer Res. 2019;38(1):1-21. doi: 10.1186/s13046-019-1131-1.
- Acar K, Uz B. A chronic myeloid leukemia case with a variant translocation t (11; 22)(q23; q11. 2): masked Philadelphia or simple variant translocation? Pan Afr Med J. 2018; 30: 1-4. doi: 10.11604/pamj.2018.30.161.9318
- Kang ZJ, Liu YF, Xu LZ, Long ZJ, Huang D, Yang Y, et al. The Philadelphia chromosome in leukemogenesis. Chin J Cancer. 2016;35(1):1-15. doi: 10.1186/s40880-016-0108-0
- Piedimonte M, Ottone T, Alfonso V, Ferrari A, Conte E, Divona M, et al. A rare BCR-ABL1 transcript in Philadelphia-positive acute myeloid leukemia: case report and literature review. BMC Cancer. 2019;19(1):1-6. doi: 10.1186/s12885-019-5265-5
- Reckel S, Hamelin R, Georgeon S, Armand F, Jolliet Q, Chiappe D, et al. Differential signaling networks of Bcr–Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia. 2017;31(7):1502-1512. doi: 10.1038/leu.2017.36
- Hakeem A, Shiekh AA, Bhat GM, Lone AR. Prognostification of ALL by Cytogenetics. Indian J Hematol Blood Transfus. 2015;31(3):322-331. doi: 10.1007/s12288-014-0483-0
- Ravandi F, Kebriaei P. Philadelphia chromosome-positive acute lymphoblastic leukemia. Hematol Oncol Clin North Am. 2009;23(5):1043-1063. doi: 10.1016/j.hoc.2009.07.007
- Landstrom AP, Tefferi A. Fluorescent in situ hybridization in the diagnosis, prognosis, and treatment monitoring of chronic myeloid leukemia. Leuk Lymphoma. 2006;47(3):397-402. doi: 10.1080/10428190500353133
- Sugapriya D, Preethi S, Shanthi P, Chandra N, Jeyaraman G, Sachdanandam P, et al. BCR-ABL translocation in pediatric acute lymphoblastic leukemia in Southern India. Indian J Hematol Blood Transfus. 2012;28(1):37-41. doi: 10.1007/s12288-011-0096-9
- Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome–like acute lymphoblastic leukemia. Blood. 2017;130(19):2064-2072. doi: 10.1182/blood-2017-06-743252
- Rowly JD. A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290-293. doi: 10.1038/243290a0
- Kurzrock R, Gutterman JU, Talpaz M. The molecular genetics of Philadelphia chromosome-positive leukemias. N Engl J Med. 1988;319(15):990-998. doi: 10.1056/NEJM198810133191506
- Li S, Ilaria Jr RL, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia–like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399-1412. doi: 10.1084/jem.189.9.1399
- Fernandes MS, Reddy MM, Gonneville JR, DeRoo SC, Podar K, Griffin JD, et al. BCR-ABL promotes the frequency of mutagenic single-strand annealing DNA repair. Blood. 2009;114(9):1813-1819. doi: 10.1182/blood-2008-07-172148
- Sattler M, Griffin JD. Mechanisms of transformation by the BCR/ABL oncogene. Int J Hematol. 2001;73(3):278-291. doi: 10.1007/BF02981952
- Stock W. Advances in the treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Clin Adv Hematol Oncol. 2008;6(7):487-488.
- Awan T, Iqbal Z, Aleem A, Sabir N, Absar M, Rasool M, et al. Five most common prognostically important fusion oncogenes are detected in the majority of Pakistani pediatric acute lymphoblastic leukemia patients and are strongly associated with disease biology and treatment outcome. Asian Pac J Cancer Prev. 2012;13(11): 5469-5475. doi: 10.7314/APJCP.2012.13.11.5469
- Faiz M, Qureshi A, Qazi JI. Molecular characterization of different fusion oncogenes associated with childhood acute lymphoblastic leukemia from Pakistan. Int J Agro Vet Med Sci. 2011;5(5)497-507.
- Siddiqui R, Nancy N, Naing WP, Ali S, Dar L, Khan BK, et al. Distribution of common genetic subgroups in childhood acute lymphoblastic leukemia in four developing countries. Cancer Genet Cytogenet. 2010;200(2):149-153. doi: 10.1016/j.cancergencyto.2010.04.010
- Pandita A, Harish R, Digra SK, Raina A, Sharma AA, Koul A. Molecular cytogenetics in childhood acute lymphoblastic leukemia: A hospital-based observational study. Clin Med Insights Oncol. 2015;9:39-42. doi: 10.4137/CMO.S24463
- Schlieben S, Borkhardt A, Reinisch I, Ritterbach J, Janssen JW, Ratei R, et al. Incidence and clinical outcome of children with BCR/ABL-positive acute lymphoblastic leukemia (ALL). A prospective RT-PCR study based on 673 patients enrolled in the German pediatric multicenter therapy trials ALL-BFM-90 and CoALL-05-92. Leukemia. 1996;10(6):957-963.
- Lenk L, Alsadeq A, Schewe DM. Involvement of the central nervous system in acute lymphoblastic leukemia: opinions on molecular mechanisms and clinical implications based on recent data. Cancer Metastasis Rev. 2020;39(1):173-187. doi: 10.1007/s10555-020-09848-z
- Cao P, Yu Y, Wang W, Xu H, He Y. Fluorescence in situ hybridization comparison of the prognostic factors in adult and pediatric acute lymphoblastic leukemia: A retrospective analysis of 282 cases. Exp Ther Med. 2018;16(6):4674-4684. doi: 10.3892/etm.2018.6821
- Artigas AC, Cabrera CM, Melo AA, Páez FE, Arriagada MM, Astete AC, et al. Frequency of TEL/AML1 and BCR/ABL fusion genes in children with acute lymphoblastic leukemia. Rev Med Chil. 2006;134(11):1367-1376. doi: 10.4067/s0034-98872006001100003
- Trka J, Zuna J, Haskovec C, Brabencova A, Kalinova M, Muzikova K, et al. Detection of BCR/ABL, MLL/AF4 and TEL/AML1 hybrid genes and monitoring of minimal residual disease in pediatric patients with acute lymphoblastic leukemia. Cas Lek Cesk. 1999;138(1):12-17.
- Siraj AK, Kamat S, Gutierrez MI, Banavali S, Timpson G, Sazawal S, et al. Frequencies of the major subgroups of precursor B-cell acute lymphoblastic leukemia in Indian children differ from the West. Leukemia. 2003;17(6):1192-1193. doi: 10.1038/sj.leu.2402931
- Bekker-Méndez VC, Miranda-Peralta E, Núñez-Enríquez JC, Olarte-Carrillo I, Guerra-Castillo FX, Pompa-Mera EN, et al. Prevalence of gene rearrangements in Mexican children with acute lymphoblastic leukemia: a population study—Report from the Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia. Biomed Res Int. 2014;2014:1-9. doi: 10.1155/2014/210560
This is an open-access article distributed under the terms of the CreativeCommons Attribution License (CC BY) 4.0 https://creativecommons.org/licenses/by/4.0/
