By Waqas Ahmad1, Sara Sadiq2, Ambreen Barkat3, Muhammad Fazal Hussain Qureshi4
AFFILIATIONS:
- Institute of Biochemistry and Biotechnology, University of Gujrat.
- Department of Physiology, CMH Institute of Medical Sciences, Bahawalpur.
- Department of Biochemistry, Pir Mehr Ali Shah Arid Agriculture University, Rawalpindi.
- Student, Ziauddin University Karachi, Pakistan.
ABSTRACT
Background: Psoriasis Vulgaris (PsV) is one of the most severe chronic, immune mediated skin diseases. The Notch signaling pathway, a key regulator for epidermal renewal, contributes in differentiation, proliferation and survival of keratinocytes. Alterations in the NOTCH4 gene disrupts Notch pathway. The current study aimed to find out the association of NOTCH4 gene polymorphism(s) in Pakistani psoriatic patients.
Methods: A case control study, 390 DNA samples (190 samples of Psoriasis Vulgaris and 200 healthy control individuals), from January-December 2019, were selected from Rawalpindi Leprosy Hospital and healthy population, respectively. For amplification of (rs387071) SNP of NOTCH4 gene, lab standard protocols for T-ARMS-PCR were followed. Frequencies of genotype and allele were calculated by using Hardy Weinberg theorem. Data were analyzed by using SPSS and p-value ≤0.05 was considered statistically significant.
Results: The mean age of patients was 34.6±14 while among control subjects it was 32.8±10.0. In cases, genotype homozygous A/A119 (62.6%) was more prevalent, followed by heterozygous A/G171 (90%) while homozygous G/G19 (10%) was the least prevalent between cases and controls. Allele A frequency in diseased subjects was 0.76 while, for controls it was 0.78. In addition, allele G frequency in patients and controls was 0.24 and 0.22 respectively. Based on Hardy Weinberg equilibrium, no association of (rs387071) NOTCH4 gene with psoriasis cases was found.
Conclusion: NOTCH4 gene (rs387071) polymorphism was not significantly associated with patients of psoriasis Vulgaris in Pakistan. Larger studies are required to establish ethnic-specific markers for psoriasis in Pakistani population.
Keywords: Psoriasis Vulgaris; PCR; NOTCH4; Polymorphism.
Psoriasis Vulgaris (PsV) is one of the most severe chronic, immune mediated skin diseases, with the high rate of morbidity and mortality worldwide. The worldwide prevalence of psoriasis is ranging from 0.91% up to 8.5% and the majority of the cases are from developing countries. Its frequency depends upon the age, ethnicity and geographic variations that are high in the regions away from the equator. Approximately two-thirds of people with psoriasis have a mild form of this disease, with less than 3% of the cutaneous surface of the body affected, but others have the widespread contribution of the skin affected1,2.
It is a characteristically lifelong disorder. There is no medication for the disease, but its symptoms can be minimized by different treatments. It has five main types, which are inverse, guttate, plaque, erythrodermic and pustular. Out of these types, the most prevalent is plaque psoriasis, in which white and red shades of scaly lesions produce on epidermal layer. These lesions are produced by epidermal maturation and hyper proliferation3,4. The immune system plays a main role in the pathophysiology of psoriasis. Despite past researches looking at many causes, the master switch is still unknown that turns on psoriasis. It is believed that in Psoriasis, T-cell travels to the dermis of the skin and discharge cytokines (TNFα) that cause infection and greater assembly of skin cells. There are multiple etiological factors including genetic and environmental factors. Numerous genes and single nucleotide polymorphisms (SNPs) play a starring role in the development of psoriasis5-7.
The Notch signaling pathway acts as a key regulator for several critical aspects of epidermal renewal, such as differentiation, proliferation and survival of keratinocytes8-10. In humans, four NOTCH signaling transmembrane receptors are NOTCH 1-4, that bind to ligands Delta 1-3and Jagged 1-2 at neighboring cells8,11. Alterations in the Notch pathway disrupt its functions e.g. the keratinocyte differentiation program, which leads to disorganization of the entire epidermal multi-stratum structure, the dermoepidermal junction and dermal invasion8,12. Moreover, it results in the histogenesis of neoplasm and in the progress of psoriasis13,14. Keeping in view, the role of NOTCH4 gene in psoriasis pathogenesis, the current study aimed to find out the association of NOTCH4 gene polymorphism(s) in Pakistani psoriatic patients.
A case control study was conducted for which cases were selected from Rawalpindi Leprosy Hospital while controls were selected from the healthy general population. All cases were diagnosed patients of Psoriasis Vulgaris (PsV). Patients with the associated disease were not included in the study, to avoid possible interfaces with potentially coexisting genetically determined conditions. Healthy subjects, with no psoriasis disease, and in the same sex and age group were enrolled as controls. All the Psoriasis patients were investigated based on an in-depth performa, containing all demographic information. The written informed consent form was also presented to attain acceptance from patients. The study got approval from the Ethics Committee of PMAS–Arid Agriculture University Rawalpindi for the use of human subjects with a reference Number: ABPA00129.
Blood samples were taken from both controls and Psoriasis patients. About 4-5ml venous blood was drawn by using syringes. Samples were collected in EDTA vacutainers and DNA extraction was performed as soon as the blood samples were taken to avoid DNA denaturation. From whole blood, DNA was extracted by using a revised protocol15. 750µl blood was mixed with 750µl of Red Blood Cell (RBC) Lysis buffer, and then at room temperature incubated for 10 min. Samples were then centrifuged at 12000 rpm for 10 minutes until the WBCs pellet was obtained. For proper RBC lyses, this step was repeated 3,4 times, supernatant was discarded, when WBC pellet formed. 450µl Nuclear Lysis Buffer and 40µl of 10% SDS was added into pellet. Samples were protected at 37°C for overnight. 300µl of phenol and 300µl T.E buffer were added and then centrifuged for 15 minutes at 10000 rpm. The upper layer in a tube was collected in a 1.5ml tube with the same amount of chloroform; isoamyl alcohol (24:1) was transferred in the supernatant and then centrifuged at 12000 rpm for 15 minutes. After centrifugation, the supernatant was again collected in a new tube and 90µl of 3M CH3COONa and 600µl isopropanol was added. DNA precipitated out when the tube was gently upturned for few minutes and again centrifugation at 12000 rpm for 15 min was done. DNA palette was settled and then washed with 70% ethanol. In order to evaporate the remaining ethanol, tubes were left to dry, and 100µl TE buffer was added to dissolve the DNA palette.
Extracted DNA was stored at -20ºC until further use. The presence of genomic DNA stocks was confirmed by using 1% agarose gel. The size of sample DNA was compared with the DNA ladder (Fermentas) of 100 bp and 50bp. To examine the gel images, Gel documentation system (Alpha Innotech) was used for record and results in analysis. SNP (rs387071) sequence of NOTCH4 gene was retrieved from NCBI dbSNP databases. Primer designing was done by using the online web resource Primer 3 program. The sequence used was: F(O):CTGTATGCTTAAAAATGCCAGTATCGGC;R(O):TTTTTTTTTTTTGAGACAGTCCAGGCTG;F(G):TGTCTCTACTAAAATACAAAAAGTCAGACG;R(A):GATTACAGGCGCCCACCACCACGACT. For amplification of (rs387071) SNP of NOTCH4 gene, lab standard protocols for tetra-primer amplification refractory mutation system PCR (T- ARMS PCR) were followed. Before the amplification step, the stock genomic DNA and dilutions were run on 0.8% and 1% agarose gel, and quantification was done by using NanoDrop (Avans, AUVS-201) to estimate the quantity and quality of stock and diluted DNA suitable for PCR reaction. DNA was amplified by initial denaturation at 95°C for 5 minutes and then 35 cycles comprising of denaturation at 95°C for 40 seconds, annealing at 59.4°C for 40 seconds and extension for 1 minute at 72°C. The final extension was carried out at 72°C for 10 minutes.
Genotyping of all subjects was acquired by direct picturing of bands on the gel. Depending on genotypes, frequencies of genotype and allele were calculated by using Hardy Weinberg theorem15. Data were analyzed by using SPSS version-20. Mean and standard deviation were calculated for numerical data while frequency and percentages for qualitative data. For genotype data, Hardy Weinberg equilibrium was applied. Odds ratios were calculated to find association of genotype among cases and controls, p-value ≤0.05 was considered as statistically significant.
The mean age of patients was 34.6±14 in cases and 32.8±10.0 in controls. Regarding the age of onset, patients were categorized as early-onset if they were first detected before the age of 40 years and late onset if they were diagnosed after 40 years. In cases, 63.9% were early-onset and 27.3% were late-onset with male predominance in both groups. About 4.2% of the participants had a positive family history of psoriasis. Based on body surface area affected among studied cases, most cases were those in which disease was diagnosed 4-5 years ago and they had a severe form of psoriasis, and their whole body was affected.
The 390 DNA samples (190 samples of Psoriasis Vulgaris and 200 healthy control individuals) were genotyped for SNP (rs387071) of NOTCH4 gene polymorphism. To count genotypes, direct gene counting was used. Gel analyses were done for polymorphism profile and also to find the number of subjects that belong to a certain type of genotype. Polymorphism profile was made by comparing gel images with 100bp and 50bp markers. Out of different T- ARMS PCR products, the 11- 25 products were shown at 50 bp DNA ladder in Figure 1.

Figure 1: Tetra-primer amplification refractory mutation system-polymerase chain (T-ARMS-PCR) for single-nucleotide polymorphisms (SNPs) (rs387071). Comparison with ladder reported that samples 16, 17, 21, 22, 23, 24, 25 were homozygous for genotype AA while, 14, 18 were homozygous for genotype GG.
Hardy Weinberg principle was tested for gene frequencies calculation. In cases, the genotype frequencies calculated for all three genotypes were 0.63 A/A, 0.27 A/G and 0.1 G/G while, in controls genotype frequencies were 0.64 for A/A, 0.28 for A/G and for G/G 0.08 respectively. No significant difference between these two groups was found. In cases, genotype homozygous A/A was more prevalent genotype, followed by heterozygous A/G while homozygous G/G was the least prevalent between cases and controls as shown in Figure 2.
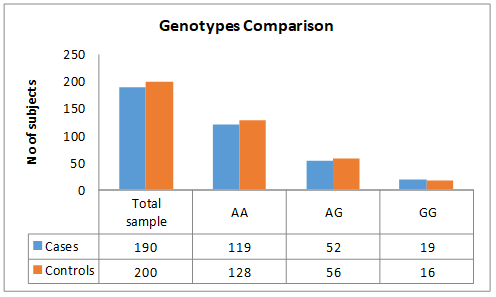
Figure 2: Bar chart showing the comparison of genotypes between cases and controls.
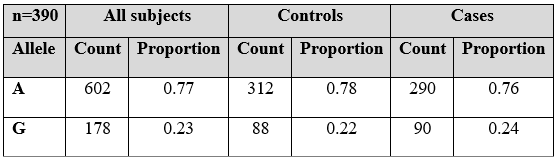
Allele A frequency in diseased subjects was 0.76 while, for controls it was 0.78. Moreover, allele G frequency in patients and controls was 0.24 and 0.22 respectively as mentioned in Table 1. NOTCH4gene (rs387071) was genotyped but based on Hardy Weinberg equilibrium there was no association of (rs387071) NOTCH 4 gene with psoriasis cases.
Table 1: Allelic frequency of (rs387071) in psoriatic patients.
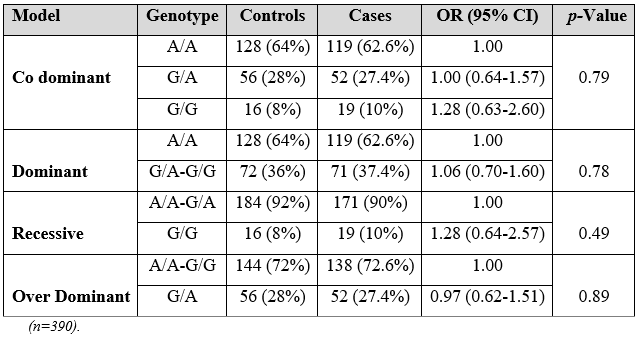
As Table 2 reported non-significant allelic and genotypic difference was found among patients and controls and the p-values were non-significant.
Table 2: Genotype exact test for Hardy-Weinberg Equilibrium.
Psoriasis can begin at any age, so on the basis of age of onset, Psoriasis has been categorized in two types, one is patients with early-onset or Type I psoriasis (before the age of 40). These persons suffer from more severe disease with a family history. Patients with late-onset or Type II psoriasis (after the age of 40) have a milder disease16. The current study favored this finding by reporting 63.9% cases with early-onset and 27.3% with late-onset while only 4.2% of the participants had a positive family history of psoriasis. Di Meglio et al. reported that the pathogenesis of psoriasis results due to complex interface among genetic, immunological, and environmental factors17. Looking over the genetic analysis, classic genome wide linkage analysis has recognized nine loci on different chromosomes related to psoriasis, named as the psoriasis susceptibility 1 through 9 (PSORS1 to PSORS9). Certain variations in these genes are found in psoriasis. The main determinant is PSORS1, which possibly accounts for 35-50% of its heritability. It controls genes that affect the immune system or encrypt proteins that are found on the skin surface in more amounts in psoriasis. PSORS1 is located on chromosome 6 in the MHC region, which switches important immune roles. Three genes in the PSORS1 locus have a robust relationship with psoriasis18,19.
Literature reported that NOTCH4 gene deviations could be intricate in the clinical appearance of psoriasis which specifies that VEGF-induced NOTCH4 over manifestation stimulates endothelial cell morphology alterations and also increased dermis vessel permeability that continues to the development of psoriatic abrasions14. TheNOTCH4 gene is recognized as INT3, located on 6p21.3. Kourmouli et al. investigated the association of polymorphisms NOTCH4 gene (rs387071) with PsV. They studied 305 patients with Psoriasis Vulgaris, genotyped by real time quantitative PCR or PCR-RFLP. Results of their study reported that NOTCH4 polymorphisms appear to play an indubitable role in epidermal differentiation and keratinization disturbances in psoriasis patients, due to the significant association of relevant polymorphisms (rs387071) with the clinical expression of this disease. In psoriasis, angiogenesis begins before visible epidermal hyperplasia and may be an inducer of its development14,20,21. However, the current study neglected the role of NOTCH4 gene (rs387071) polymorphism, as no significant difference was found between cases and controls. The reason behind this variation might be either ethnicity or geographical variations. Another study was done in Italy support the current finding by reporting the expression of NOTCH1 and NOTCH2 in their psoriatic population, instead of NOTCH 4 gene expression22.
Agha et al. studied psoriasis susceptibility association, earlier stated in Chinese and Caucasian populations in a Pakistani cohort. Results of their study showed that HLA-Cw6 has the strongest association and signifying that the HLA-Cw1-B46 risk haplotype and may exist in the Pakistani population. Their study concluded that HLA-Cw6 is related to psoriasis predisposition in the Pakistani population, formerly reported in other populations19,23,24.
Looking over the SNPs, studies reported a higher frequency of G allele and the genotype observed was G/G followed by A/G (14) but the current study contradicts this finding by reporting a higher frequency of A allele with A/A genotype. It had also been reported that the majority of the cases were in the age group of late-onset and were female14,25 while the current study supported this finding but the majority affected were male. The gender-based variation in the occurrence of the disease might be due to the involvement of sex hormones in the pathogenesis of PsV. NOTCH4 receptor cross talk with the estrogen receptors thus regulating the Notch signaling pathway in endothelial cells26.
NOTCH4 gene (rs387071) polymorphism marker was not found significant among the patients of Psoriasis Vulgaris in Pakistan. However, investigation with large study samples might be helpful in exploring the potential genes contributing for Pakistani population.
We like to acknowledge and extend our gratitude to Rawalpindi Leprosy Hospital and all the participants who participated in this research.
The authors declare no conflict of interest.
The study received approval from the Ethics Committee of PMAS-Arid Agriculture University Rawalpindi for the use of human subjects (Reference Number: ABPA00129).
Patient consents were obtained before sample collection.
All authors contributed equally in this study. All authors approved the final version of the manuscript.
- Rachakonda TD, Schupp CW, Armstrong AW. Psoriasis prevalence among adults in the United States. J Am Acad Dermatol. 2014;70(3):512-516.
- Parisi R, Symmons DP, Griffiths CE, Ashcroft DM. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol. 2013;133(2):377-385.
- Zangeneh FZ, Shooshtary FS. Psoriasis—types, causes and medication [Internet]. In psoriasis-types, causes and medication: IntechOpen; 2013 [cited 2020 Dec 28]. Aavailble from: https://www.intechopen.com/books/psoriasis-types-causes-and-medication/psoriasis-types-causes-and-medication
- Lima H, editor. Psoriasis: Types, Causes and Medication. BoD–Books on Demand; 2013. 3 p.
- Al-Shobaili HA, Qureshi MG. Pathophysiology of psoriasis: Current concepts. InTech; 2013. Chapter 1, Psoriasis-types, causes and medication; p. 3-38.
- Fallen RS, Mitra A, Morrisey L, Lima H. Pathophysiology of psoriasis: Current concepts BoD – Books on Demand. Copyright. 2013. Chapter 3, Psoriasis as a chess board—An update of psoriasis pathophysiology; p. 57-90.
- McKenzie RC, Sabin E. Aberrant signalling and transcription factor activation as an explanation for the defective growth control and differentiation of keratinocytes in psoriasis: a hypothesis. Exp Dermatol. 2003;12(4):337-45.
- Okuyama R, Tagami H, Aiba S. Notch signaling: its role in epidermal homeostasis and in the pathogenesis of skin diseases. J Dermatolo Sci. 2008;49(3):187-194.
- Panelos J, Massi D. Emerging role of Notch signaling in epidermal differentiation and skin cancer. Cancer Biol Ther. 2009;8(21):1986-1993.
- Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol. 2008;20(2):171-179.
- Thélu J, Rossio P, Favier B. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2002;2(1):1-12.
- Rooney P, Connolly M, Gao W, McCormick J, Biniecka M, Sullivan O, et al. Notch-1 mediates endothelial cell activation and invasion in psoriasis. Experimental Dermatol. 2014;23(2):113-118.
- Abdou AG, Maraee AH, Sharaf A, Elnaidany NF. Up-regulation of Notch-1 in psoriasis: an immunohistochemical study. Ann Diagn Pathol. 2012;16(3):177-184.
- Kourmouli N, Tripsianis G, Diplas A, Veletza S. Notch2, notch4 gene polymorphisms in psoriasis vulgaris. Eur J Dermatol. 2013;23(2):146-153.
- Hu X, Duan X, Pan D, Zhang S, Li Q. A model-embedded trend test with incorporating Hardy-Weinberg equilibrium information. J Syst Sci Complex. 2017;30(1):101-110.
- Langley R, Krueger G, Griffiths C. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis. 2005;64(Suppl 2):18-23.
- Di Meglio P, Perera GK, Nestle FO. The multitasking organ: recent insights into skin immune function. Immun. 2011;35(6):857-869.
- Zhao M, Lu Q. The aberrant epigenetic modifications in the pathogenesis of psoriasis. J Investig Dermatol Symp Proc. 2018; 19(2):S81-S82.
- Zhu KJ, Lv YM, Yin XY, Wang ZX, Sun LD, He SM, et al. Psoriasis regression analysis of MHC loci identifies shared genetic variants with vitiligo. PLoS One. 2011;6(11):1-7.
- Pan M, Huang Y, Zhu X, Lin X, Luo D. miR‑125b‑mediated regulation of cell proliferation through the Jagged‑1/Notch signaling pathway by inhibiting BRD4 expression in psoriasis. Mol Med Rep. 2019;19(6):5227-5236.
- Marasca C, Scala E, Di Caprio R, Raimondo A, Cacciapuoti S, Balato A, et al. Notch dysregulation and hidradenitis suppurativa, psoriasis, atopic dermatitis and lichen planus: let’s talk about Numb. Br J Dermatol. 2019;180(4):950-951.
- Skarmoutsou E, Trovato C, Granata M, Rossi GA, Mosca A, Longo V, et al. Biological therapy induces expression changes in Notch pathway in psoriasis. Arch Dermatol Res. 2015;307(10):863-873.
- Agha Z, Shaiq PA, Ahmed S, Ali L, Azam M, Ali SH, Kaukab G, Qamar R. A study of ACE, eNOS and MTHFR association with psoriasis in Pakistani population. Meta Gene. 2018;15:65-69.
- Feng BJ, Sun LD, Soltani-Arabshahi R, Bowcock AM, Nair RP, Stuart P, et al. Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet. 2009;5(8):1-8.
- Ota T, Takekoshi S, Takagi T, Kitatani K, Toriumi K, Kojima T, et al. Notch signaling may be involved in the abnormal differentiation of epidermal keratinocytes in psoriasis. Acta Histochem Cytochem. 2014;47(4):175-83.
- Icen M, Crowson CS, McEvoy MT, Dann FJ, Gabriel SE, Kremers HM. Trends in incidence of adult-onset psoriasis over three decades: a population-based study. J Am Acad Dermatol. 2009;60(3):394-401.
This is an open-access article distributed under the terms of the CreativeCommons Attribution License (CC BY) 4.0 https://creativecommons.org/licenses/by/4.0/
