By Sadaf Saleem Uppal1, Bushra Chaudhry2, Abdul Khaliq Naveed3
AFFLIATIONS:
- Department of Biochemistry, Shalamar Medical and Dental College, Lahore, Pakistan.
- Department of Biological Sciences, Agha Khan Medical College, Karachi, Pakistan.
- Department of Biochemistry, Wah Medical College, Wah Cantt, Rawalpindi, Punjab, Pakistan.
DOI: https://doi.org/10.36283/PJMD11-4/003
How to cite: Uppal SS, Chaudhry B, Naveed AK. “Regenerating” Iα Gene Variants g. -385T>C and g. -243T>G in Type 2 Diabetes. Pak J Med Dent. 2022;11(4): 15-21. doi: 10.36283/PJMD11-4/003
Background: Restoration of the body’s ability to produce insulin by regeneration of β-cells is an essential step toward the management of diabetes. REG Iα is released from damaged β-cell and facilitates their renewal. However, genetic variants of REG Iα and their clinical associations are overlooked areas. This study was designed to identify polymorphisms of REG Iα gene exon 1 and its association with type 2 diabetes.
Methods: This was a case-control study conducted on individuals n=51 both diabetics (n=36) (aged, 52±9 years; males 25, females 11) (group II) and age-related controls (n=15) (aged, 50±3 years; males 9, female 6) (group I) recruited from PNS Shifa Hospital, Karachi. The samples were amplified, and the gene was sequenced to identify polymorphisms. Chi-square (γ2) test was used to determine whether each polymorphism obeys Hardy-Weinberg equilibrium (HWE). Odds ratios, 95% confidence interval along with corresponding p values were calculated through logistic regression analysis. p < 0.05 was considered statistically significant.
Results: A set of two single nucleotide polymorphisms (SNPs) were detected in the REG Iα gene exon1 in the subjects under study. SNPs studied were: g.-243T>G [rs 283890], and g.-385T>C [rs10165462]). Both variants g-243T>G, and g-385T>C showed significant association (γ2=12.8, p=0.0003, γ2=13.8, p=0.0002) with smoking. The allele G showed an eight-fold increased risk of disease in smokers while allele C had less chance of the disease in smokers as compared to non-smokers.
Conclusion: Among the SNPs detected in exon 1 of the gene for REG Iα, the g. -385T>C and g. -243T>G variants showed an association with smoking in type 2 diabetes and the SNP -243G (rs 283890) was found to increase the risk of disease in smokers.
Keywords: REG Iα gene; Polymorphisms; Type 2 Diabetes; β-cells Regeneration.
Diabetes management is a continuous challenge for medical science. Researchers around the world are striving hard to discover novel molecular markers that would carry the capability of increasing long-term survival as well as functional efficacy of β-cells1,2. It is suggested that human insulin-secreting cells can proliferate and induce the synthesis of several progenitor/stem cells. Many genes and transcription factors take part in this process like Reg islets derived proteins, NeuroD1, Sox 9, Netrin 1, and Neurogenin-3 3. In humans, REG gene family members have been reported and some of them have been associated with regeneration and neogenesis of β-cells4. The REG gene expression in the pancreas is induced during morphogenesis of islets or by damage to β-cells5,6. It rejuvenates the β-cell population and improves experimentally induced diabetes7,8. The β-cell DNA replication occurs in an autocrine or paracrine mode. A supposed Reg protein receptor was recognized that transmits the signals for β-cells growth and regeneration9.
Reg I the first member of the family is produced by exocrine pancreas acinar cells and resides in β-cells secretory granules along with insulin5. REG Iα is encoded by an approximately 2.7 kb gene, comprising six exons and five intervening sequences located on chromosome 2p12. Expression of the REG I gene has been observed early in pancreas development and islet cell expansion. Reduced levels of Reg I protein and insulin were observed in the case of aging and pancreatitis-induced diabetes10. Lately, a study suggested that damage to acinar cells is related to the insufficiency of insulin in chronic pancreatitis. Based on the review of studies, it is proposed that lost defense from acinar cells of exocrine pancreas secreting regenerating proteins results in damage of insulin-secreting β-cells in chronic pancreatitis11.
Reg proteins upregulation in individuals with type 2 diabetes after gastric bypass surgery may contribute to its remission12. Furthermore, Reg I overexpressed after virus infection induced diabetes in REG I knockout mice specifies their crucial role in beta cell regeneration13. Raised levels of Reg Iα and Iβ demonstrated a better survival rate in pancreatic cancer14. Studies are being done to identify miRNAs targeting human Reg family members that may provide new insights into Reg protein biology15.
However, genetic variants of REG Iα and their clinical associations are overlooked areas that need to be explored. A study carried out to screen the comprehensive REG Iα gene showed no association between REG I gene abnormalities and diabetes. Korean population research for genetic variants in REG Iα found some of the polymorphisms showed modest associations with type 2 diabetes of early onset16. Moreover, risk factors associated with diabetes like BMI, family history of the disease, age, sex, smoking and hypertension and their association with the genetic variants of REG Iα need to be explored. To meticulously explore the possible REG Iα gene link with type 2 diabetes, the study aimed to identify polymorphisms of REG Iα gene exon 1 variant in Pakistani targeted people and their possible link with type 2 diabetes and associated risk factors.
A total of n=51 participants were included in this case-control study. The type 2 diabetes n=36 patients (aged, 52+9 years; males 25, females 11) were recruited from PNS Shifa Hospital, Karachi medical OPD, and n=15 healthy controls (aged, 50+3 years; males 9, female 6). The study was approved by the Ethical Review Committee of the Centre for Research in Experimental and Applied Medicine (CREAM), Army Medical College, (Letter No. 02/CREAM-A/Sadaf Saleem). The patients with type 2 diabetes diagnosed under the American Diabetes Association (ADA) criteria were selected. The participants were well-versed in the study purpose and written informed consent was taken before preceding sampling.
Blood samples were drawn under an aseptic technique for biochemical measurements and DNA extraction. Whole blood was collected in EDTA tubes and DNA extraction was done using a modified Sambrook and Russell 2001 methodology. Primers were designed for exon 1 and their corresponding intronic regions using Primer-BLAST software as follows: FORWARD PRIMER: TCCCAAAACTCACCGCTTGC, and REVERSE PRIMER: CTCTGAGACACCCACACCTTC. The primers amplified the region of 322 bp.
All samples were amplified using primer set for exon 1 after its optimization on Bioer’s XP PCR Thermal cycler (Pre denaturation at 94⁰C for 4 minutes, denaturation at 94⁰C for 30 seconds, annealing at 55⁰C for 40 seconds, extension at 72⁰C 1kb/min for 35 cycles, post extension 72⁰C for 10 minutes, hold 4⁰C. PCR products amplified were run on agarose gel (2%) and visualized on GelDoc imaging system using Herolab E.A.S.Y Win 32 program. PCR products were purified through a PCR cleanup kit using Spin Column Method. PCR product quantification was done by Spectrophotometer using 260 nm UV absorbance. Sanger dideoxy method was used to sequence DNA of interest using “BigDye Terminator v 3.1 cycle sequencing kit” on Genetic Analyzer.
Sequence Scanner 2 and Clustal X software were used to analyze the chromatograms and alignment between sequences, respectively. Alignment between REG Iα gene sequence of all the study population and known sequence from the National Center for Biotechnology Information (NCBI) was done using Nucleotide-BLAST and SNPs were identified in both the type 2 diabetes group and controls. Known polymorphisms for the gene of interest from the Gene Card database and dbSNP NCBI Nucleotide BLAST were used for comparison with the newly identified SNPs.
To analyze all the data SPSS statistical software was used. Descriptive statistics were calculated for clinicodemographic and anthropometric data. The normality of the data was checked by applying the Shapiro-Wilk test. Mean ± SD and Median (IQR) for non-parametric data were calculated. An Independent sample t-test was applied and p < 0.05 was considered significant and p < 0.001 highly significant. Chi-square (γ2) test was used to determine whether each polymorphism obeys Hardy-Weinberg equilibrium (HWE). Odds ratios, 95% confidence interval along with corresponding p values were calculated through logistic regression analysis.
A total of 36 individuals with diabetes (group II) and 15 controls (group I) were included in the present study. There were 9(60%) men and 6(40%) women as controls while individuals with type 2 diabetes included 25(69%) men and 11(31%) women. The mean age of Group I was 50+3 and group II was 52±9 (p=0.479). The Body mass index (BMI) of controls was 24.6±4, which was less than the diabetic group (26±3) of both groups. Whereas the Fasting blood glucose was also found high 8 (6.1-10.9) in dietetics than controls at 4.1 (3.7-5.6) 9, p <0.001). Table 1 shows the clinicodemographic and anthropometric data of the groups under study.
Table 1: Clinicodemographic and anthropometric characters of the control and diseased groups.
| Characteristics | Group I
(Control) (n=15) |
Group II
(Diabetes) (n=36) |
p-Value |
| Age (years) | 50±3 | 52±9 | 0.479 |
| Age with the onset of disease | – | 46±9 | – |
| Disease/ time duration | – | 6±5 | – |
| Body weight in kg | 68±8.6 | 71±11.5 | 0.315 |
| Height in inches | 66±3.3 | 65±3.5 | 0.390 |
| Body mass index
(kg/m2) |
24.6±4 | 26±3 | 0.248 |
| Systolic Blood Pressure
(mmHg) |
120 (110 – 120) | 130 (120 -140) | 0.001* |
| Diastolic Blood Pressure
(mmHg) |
80 (70 – 80) | 80 (80 – 90) | 0.007* |
| Fasting blood glucose
(mmol/l) |
4.1 (3.7 – 5.6) | 8 (6.1 – 10.9) | <0.001** |
Data is stated as Means± SD (Median (IQR) for non-parametric data). p <0.001** and p <0.05* were considered significant and highly significant respectively.
The 322 bp of exon 1 was amplified and checked on agarose gel electrophoresis (Figure 1). The Sanger sequencing results showed many SNPs but two were selected’ g.-243T>G [rs 283890], and g.-385T>C [rs10165462]) for analysis and association with different parameters (Figure 2).
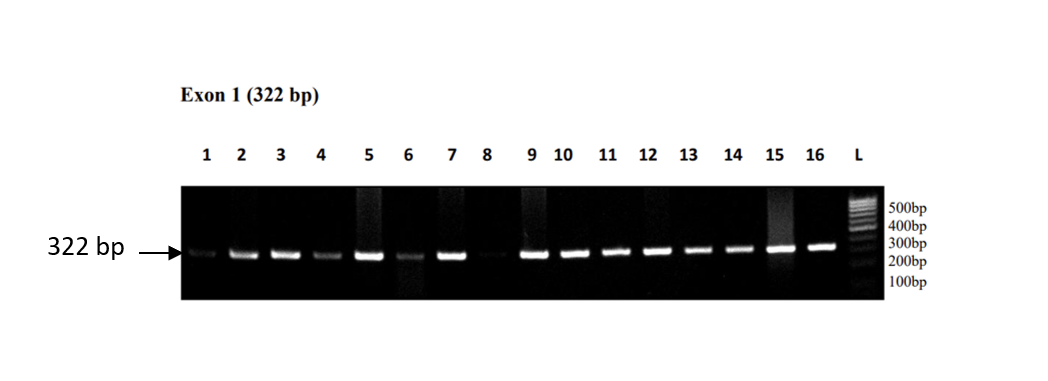
Figure 1: The 322 bp of exon 1 was amplified and checked on agarose gel electrophoresis.
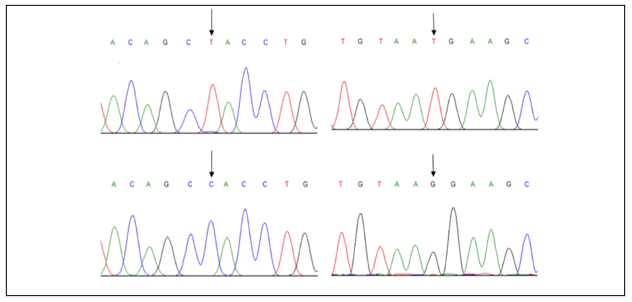
Figure 2: Left (SNP -385T>C): The electro-chromatogram of a study showing substitution of T>C (as indicated by an arrow). Right (SNP -243T>G): the electro-chromatogram of a study showing substitution of T>G (as indicated by an arrow).
Hardy Weinberg Equilibrium (HWE) was applied to determine the genotype and allelic frequencies in the SNPs -243T>G region. The selected population deviated from the HWE. For SNP2 (g.-243T>G), the OR calculated to test the relevance of frequencies of allele-T and allele-G in both controls and diabetes individuals was 0.51 with p = 0.22. However, the OR for diabetes individuals when further classified into smokers and non-smokers was 8.06 with a highly significant p<0.001 indicating an 8-fold increase in the risk of the disease in smokers (Table 2).
Table 2: Odd Ratio (OR) calculation of main variables with frequencies of alleles for SNP -243T>G.
| -243T>G | Comparative Frequencies of Cases | Total | OR/
95 % CI |
γ2 |
p-Value |
|
| Control Group | Group with Diabetes | |||||
| Allele-T | 24 | 49 | 73 | 0.53
(0.19-1.5) |
1.48 | 0.22 |
| Allele-G | 6 | 23 | 29 | |||
| 30 | 72 | 102 | ||||
| -243T>G | Male | Female | ||||
| Allele-T | 16 | 33 | 49 | 1.37
(0.45-4.2) |
0.31 | 0.57 |
| Allele-G | 6 | 17 | 23 | |||
| 22 | 50 | 72 | ||||
| -243T>G | FH (-) | FH (+) | ||||
| Allele-T | 16 | 33 | 49 | 0.90
(0.31-2.6) |
0.03 | 0.85 |
| Allele-G | 8 | 15 | 23 | |||
| 24 | 48 | 72 | ||||
| -243T>G | Age at disease onset | |||||
| < 45 | >45 | |||||
| Allele-T | 21 | 28 | 49 | 0.58
(0.20-1.6) |
1.01 | 0.31 |
| Allele-G | 7 | 16 | 23 | |||
| 28 | 44 | 72 | ||||
| -243T>G | BMI<23 | BMI>23 | ||||
| Allele-T | 9 | 40 | 49 | 0.51
(0.16-1.6) |
1.31 | 0.25 |
| Allele-G | 7 | 16 | 23 | |||
| 16 | 56 | 72 | ||||
| -243T>G | Smoker | Non-Smoker | ||||
| Allele-T | 5 | 44 | 49 | 8.06
(2.3-27.7) |
12.8 | 0.0003 |
| Allele-G | 11 | 12 | 23 | |||
| 16 | 56 | 72 | ||||
| -243T>G | Hypertension (-) | Hypertension (+) | ||||
| Allele-T | 16 | 29 | 49 | 0.89
(0.32-2.4) |
0.04 | 0.83 |
| Allele-G | 8 | 13 | 23 | |||
| 30 | 42 | 72 | ||||
For SNP1 (g.-385T>C), the OR calculated to test the relevance of frequencies of allele T and allele C in controls and diabetes individuals was OR=2.33 (0.71-7.55). The calculations were also made when diabetes individuals were further grouped. The OR calculations for smoker and non-smoker diabetes individuals was 0.11 with p<0.001 indicating that smokers with allele C have 11% less chance of the disease compared to non-smokers. For groups with age at disease onset < 45 and > 45 OR=0.32 and p = 0.06 (Table 3).
Table 3: Odd Ratio (OR) calculation of main variables with frequencies of an allele for SNP -385T>C.
| Alleles | Comparative Frequencies of Cases | Total | OR/
95 % CI |
γ2 |
p-Value |
|
| Control Group | Group with Diabetes | |||||
| -385T>C | ||||||
| Allele-T | 4 | 19 | 23 | 2.33
(0.71-7.55) |
2.06 | 0.15 |
| Allele-C | 26 | 53 | 79 | |||
| 30 | 72 | 102 | ||||
| -385T>C | Male | Female | ||||
| Allele-T | 13 | 6 | 19 | 1.06
(0.34-3.30) |
0.01 | 0.91 |
| Allele-C | 37 | 16 | 53 | |||
| 50 | 22 | 72 | ||||
| -385T>C | FH (-) | FH (+) | ||||
| Allele-T | 6 | 13 | 19 | 0.89
(0.29-2.75) |
0.03 |
0.85 |
| Allele-C | 18 | 35 | 53 | |||
| 24 | 48 | 72 | ||||
| -385T>C
|
Age at disease onset | |||||
| < 45 | >45 | |||||
| Allele-T | 4 | 15 | 19 | 0.32
(0.09-1.10) |
3.45 | 0.06 |
| Allele-C | 24 | 29 | 53 | |||
| 28 | 44 | 72 | ||||
| -385T>C | BMI<23 | BMI>23 | ||||
| Allele-T | 6 | 13 | 19 | 0.50
(0.15-1.65) |
1.30 |
0.25 |
| Allele-C | 10 | 43 | 53 | |||
| 16 | 56 | 72 | ||||
| -385T>C | Smokers | Non-Smoker | ||||
| Allele-T | 10 | 9 | 19 | 0.11
(0.03-0.39) |
13.8 |
0.0002 |
| Allele-C | 6 | 47 | 53 | |||
| 16 | 56 | 72 | ||||
| -385T>C | Hypertension (-) | Hypertension (+) | ||||
| Allele-T | 9 | 10 | 19 | 1.37
(0.47-3.94) |
0.34 |
0.55 |
| Allele-C | 21 | 32 | 53 | |||
| 30 | 42 | 72 | ||||
The study identified two SNPs in the exon 1 of the gene for REG Iα in the selected subjects and found that the variant, g.-243T>G was found to be associated with the increased risk of diabetes in smokers by eight-fold while the variants g.-385T>C was found to have a protective effect in smokers.
High values of serum Reg Iα have been reported across various types of diabetes12-15. As a marker of β-cell apoptosis and impaired kidney function16,17. Previous studies investigating the REG Iα gene in patients with fibrocalculous pancreatic diabetes and type I diabetes found no significant association between the disease and variants in the gene18,19. Wide-range scrutiny of the REG Iα gene in trophic calcific pancreatitis patients documented few polymorphisms including the one in the promoter region but did not show any disease association20. A study conducted on patients with type 2 diabetes of Korean origin found some REG Iα gene polymorphisms exhibiting associations with type 2 diabetes of early-onset nonetheless they did not find any link of these variants with general susceptibility to disease21,22.
Studies have shown that smoking causes oxidative stress and increases inflammation in the body and may lead to insulin resistance and diabetes23-25. However, not every person who smokes acquires the disease. The effect of smoking is more marked in individuals who have a genetic predisposition to diabetes. This suggests that smoking may act as the genetic modifier of the disease. Studies have shown many genetic variants associated with the risk of diabetes in smokers26,27. The results suggest that smoking may be a link between the SNP g. -243G (rs 283890) of the REG Iα gene and the development of the disease. A recent study by Azarova et al. found an association between polymorphic variant rs11927381 of IGF2BP2 gene and increased risk of type 2 diabetes therefore smoking is a foremost avertible cause of the disease28.
In addition, the hereditary variant g. -385C was found to reduce the disease risk in the age group less than 45 years. Our outcomes are in concurrence with the investigation led by the Korean populace, indicating that g.-385C brought down the risk of early-stage type 2 diabetes27. In the patients with type 2 diabetes after adjustment with numerous risk phenotypes, for example, sex, BMI, hypertension, age at disease onset, and family history, it was revealed that no polymorphism was associated significantly with the disease risk factors. Therefore, the wide-ranging quest for polymorphisms in the gene for REG Iα and the estimation of their association with type 2 diabetes are novel in Pakistani people.
A significant association between the SNP -243G (rs 283890) was found to increase the risk of disease in smokers. More studies are needed with a bigger and more diverse populace to elucidate the relationship between polymorphism in the REG Iα gene, diabetes type 2 and its related risk factors.
The authors would like to thank the entire team from CREAM lab Army Medical College, Lahore, Pathology lab, PNS Shifa hospital, Karachi and Biochemistry Research lab, Ziauddin University for providing technical support during my research work
The study was approved by the Ethical Review Committee of the Centre for Research in Experimental and Applied Medicine (CREAM), Army Medical College, (Letter No. 02/CREAM-A/Sadaf Saleem).
Informed consent was taken from patients.
SSU contributed to conception and design, drafting, acquisition of data, analysis and interpretation of data. The manuscript and revising it critically for important intellectual content. BC had made substantial contributions to the acquisition of data, analysis and interpretation of data. AKN had been involved in revising the manuscript and given final approval of the version to be published. All the authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
- Ludwig B, Barthel A, Reichel A, Block NL, Ludwig S, Schally AV, et al. Modulation of the pancreatic islet-stress axis as a novel potential therapeutic target in diabetes mellitus. Vitam Horm. 2014; 95: 195-222. doi: 10.1016/B978-0-12-800174-5.00008-9
- Chen C, Cohrs CM, Stertmann J, Bozsak R, Speier S. Human beta cell mass and function in diabetes: Recent advances in knowledge and technologies to understand disease pathogenesis. Mol Metab. 2017; 9: 943-957. doi: 10.1016/j.molmet.2017.06.019
- Bansal VS, Raja CP, Venkataraman K, Vijayalakshmi MA. Genes involved in pancreatic islet cell rejuvenation. Indian J Med Res. 2013; 137: 695-703.
- Chen Z, Downing S, Tzanakakis ES. Four decades after the discovery of regenerating islet-derived (reg) proteins: Current understanding and challenges. Front Cell Dev Biol. 2019; 7: 1-16. doi: 10.3389/fcell.2019.00235
- Li Q, Xiong X, Liu JL. The Contribution of Reg Family Proteins to Cell Growth and Survival in Pancreatic Islets. Islets of Langerhans 2nd Switzerland: Springer’s nature, 2015; 955-987. doi: 10.1007/978-94-007-6884-0_47-2
- Rankin MM, Kushner JA. Aging induces a distinct gene expression program in mouse islets. Islets. 2010; 2: 345-352. doi: 10.4161/isl.2.6.13376
- Levine JL, Patel KJ, Zheng QH, Shuldiner AR, Zenilman ME. A recombinant rat regenerating protein is mitogenic to pancreatic derived cells. J Surg Res. 2000; 89: 60-65. doi: 10.1006/jsre.1999.5800
- Luo C, Yu LT, Yang MQ, Li X, Zhang ZY, Alfred MO, et al. Recombinant Reg3β protein protects against streptozotocin-induced β-cell damage and diabetes. Sci Rep. 2016;6(1):1-13. doi: 10.1038/srep35640
- Kobayashi S, Akiyama T, Nata K, Abe M, Tajima M, Shervani NJ, et al. Identification of a receptor for reg (regenerating gene) protein, a pancreatic beta-cell regeneration factor. J Biol Chem. 2000; 275: 10723-10726. doi: 10.1074/jbc.275.15.10723
- Bluth MH, Zenilman M, inventors; Research Foundation of State University of New York, assignee. Pancreatic regenerating protein I in chronic pancreatitis and aging: implications for new therapeutic approaches to diabetes. Pancreas. 2008; 37: 386-395.
- Huan C, Stanek A, Mueller C, Ou P, Dong S, Zhang J, et al. Loss of Reg proteins’ protection of islet β cells in chronic pancreatitis: A potential mechanism for the pathogenesis of type 3c diabetes. Curr Opin Endo Metabol Res. 2019; 5: 21-28. doi: 10.1016/j.coemr.2019.02.005
- Astorri E, Guglielmi C, Bombardieri M, Alessandri C, Buzzetti R, Maggi D, et al. Circulating Reg1α proteins and autoantibodies to Reg1α proteins as biomarkers of β-cell regeneration and damage in type 1 diabetes. Horm Metab Res. 2010; 42:955-960. doi: 10.1055/s-0030-1267206
- Bacon S, Kyithar MP, Schmid J, Rizvi SR, Bonner C, Graf R, et al. Serum levels of pancreatic stone protein (PSP)/reg1A as an indicator of beta-cell apoptosis suggest an increased apoptosis rate in hepatocyte nuclear factor 1 alpha (HNF1A-MODY) carriers from the third decade of life onward. BMC Endocr Disord. 2012; 12: 1-8 doi: 10.1186/1472-6823-12-13
- Yang J, Li L, Raptis D, Li X, Li F, Chen B, et al. Pancreatic stone protein/regenerating protein (PSP/reg): a novel secreted protein up-regulated in type 2 diabetes mellitus. Endocr J. 2015; 48: 856-862. doi: 10.1007/s12020-014-0427-3
- Uppal SS, Naveed AK, Baig S, Chaudhry B. Expression of REG Iα gene in type 2 diabetics in Pakistan. Diabetol Metab Syndr. 2015;7(1):1-7. doi: 10.1186/s13098-015-0092-6
- Li L, Jia D, Graf R, Yang J. Elevated serum level of pancreatic stone protein/ regenerating protein (PSP/reg) is observed in diabetic kidney disease. Oncotarget. 2017; 13: 38145-38151. doi: 10.18632/oncotarget.16369
- Zhu H, Zhu X, Lin H, Liu D, Dai Y, Su X, et al. Association of serum PSP/REG Iα with renal function in type 2 diabetes mellitus. J Diabetes Res. 2020; 2020:1-6. doi: 10.1155/2020/9787839
- Sala P, Torrinhas RS, Fonseca DC, Heymsfield S, Giannella-Neto D, Waitzberg DL. Type 2 diabetes remission after Roux-en-Y gastric bypass: evidence for increased expression of jejunal genes encoding regenerating pancreatic islet-derived proteins as a potential mechanism. Obes Surg. 2017; 27: 1123-1127. doi: 10.1007/s11695-017-2602-0
- Aida K, Kobayashi T, Takeshita A, Jimbo E, Nishida Y, Yagihashi S, et al. Crucial role of Reg I from acinar-like cell cluster touching with islets (ATLANTIS) on mitogenesis of beta cells in EMC virus-induced diabetic mice. Biochem Biophys Res Comm. 2018; 503: 963-969. doi: 10.1016/j.bbrc.2018.06.103
- Hawrami K, Mohan V, Bone A, Hitman GA. Analysis of islet regenerating (reg) gene polymorphisms in fibrocalculous pancreatic diabetes. Pancreas. 1997; 14: 122-125.
- Koo BK, Cho YM, Kimm K, Lee JY, Oh B, Park BL, et al. Polymorphisms of the reg 1α gene and early onset type 2 diabetes in the Korean population. Korean Diabetes J. 2010;34(4):229-236. doi: 10.4093/kdj.2010.34.4.229
- Mahurkar S, Bhaskar S, Reddy DN, Rao GV, Chandak GR. Comprehensive screening for reg1α gene rules out association with tropical calcific pancreatitis. World J Gastroenterol. 2007; 13(44): 5938-5943. doi: 10.3748/wjg.v13.i44.5938
- Lee J, Taneja V, Vassallo R. Cigarette smoking and inflammation: cellular and molecular mechanisms. J Dent Res. 2012;91:142-149. doi: 10.1177/0022034511421200
- Isik B, Ceylan A, Isik R. Oxidative stress in smokers and non-smokers. Inhal Toxicol. 2007;19:767-769. doi: 10.1080/08958370701401418
- Halim M, Halim A. The effects of inflammation, aging and oxidative stress on the pathogenesis of diabetes mellitus (type 2 diabetes). Diabetes Metab Syndr. 2019;13:1165-1172. doi: 10.1016/j.dsx.2019.01.040
- Lin WY, Liu YL, Yang AC, Tsai SJ, Kuo PH. Active cigarette smoking is associated with an exacerbation of genetic susceptibility to diabetes. Diabetes. 2020;69(12):2819-2829. doi: 2337/db20-0156
- Wu P, Rybin D, Bielak LF, Feitosa MF, Franceschini N, Li Y, et al. Smoking-by-genotype interaction in type 2 diabetes risk and fasting glucose. PLoS One. 2020;15(5):1-17. doi: 10.1371/journal.pone.0230815
- Azarova IE, Klyosova EY, Lazarenko VA, Konoplya AI, Polonikov AV. rs11927381 polymorphism and type 2 diabetes mellitus: contribution of smoking to the realization of susceptibility to the disease. Bull Exp Biol Med. 2020;168: 313-316. doi: 10.1007/s10517-020-04698-9
This is an open-access article distributed under the terms of the CreativeCommons Attribution License (CC BY) 4.0 https://creativecommons.org/licenses/by/4.0/
